Comparison of Hi-C and Micro-C data. A Experimental methods of Hi
4.6 (188) · $ 11.50 · In stock

Characterizing chromatin interactions of regulatory elements and nucleosome positions, using Hi-C, Micro-C, and promoter capture Micro-C, Epigenetics & Chromatin

bin3C: exploiting Hi-C sequencing data to accurately resolve

Application of Hi-C and other omics data analysis in human cancer

Comparison of computational methods for 3D genome analysis at

An integrated model for detecting significant chromatin

figure supplement 1. Comparison of Micro-C and Hi-C mapping in
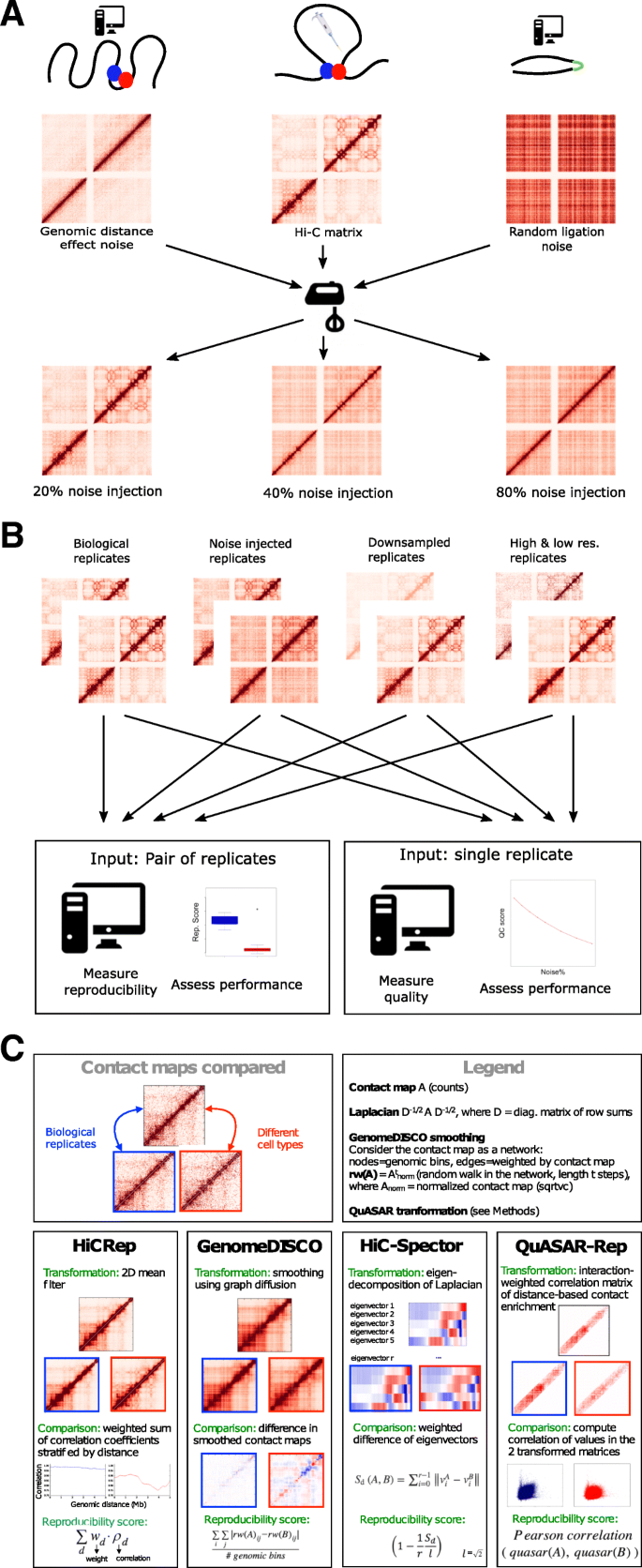
Measuring the reproducibility and quality of Hi-C data

Characterizing chromatin interactions of regulatory elements and nucleosome positions, using Hi-C, Micro-C, and promoter capture Micro-C, Epigenetics & Chromatin
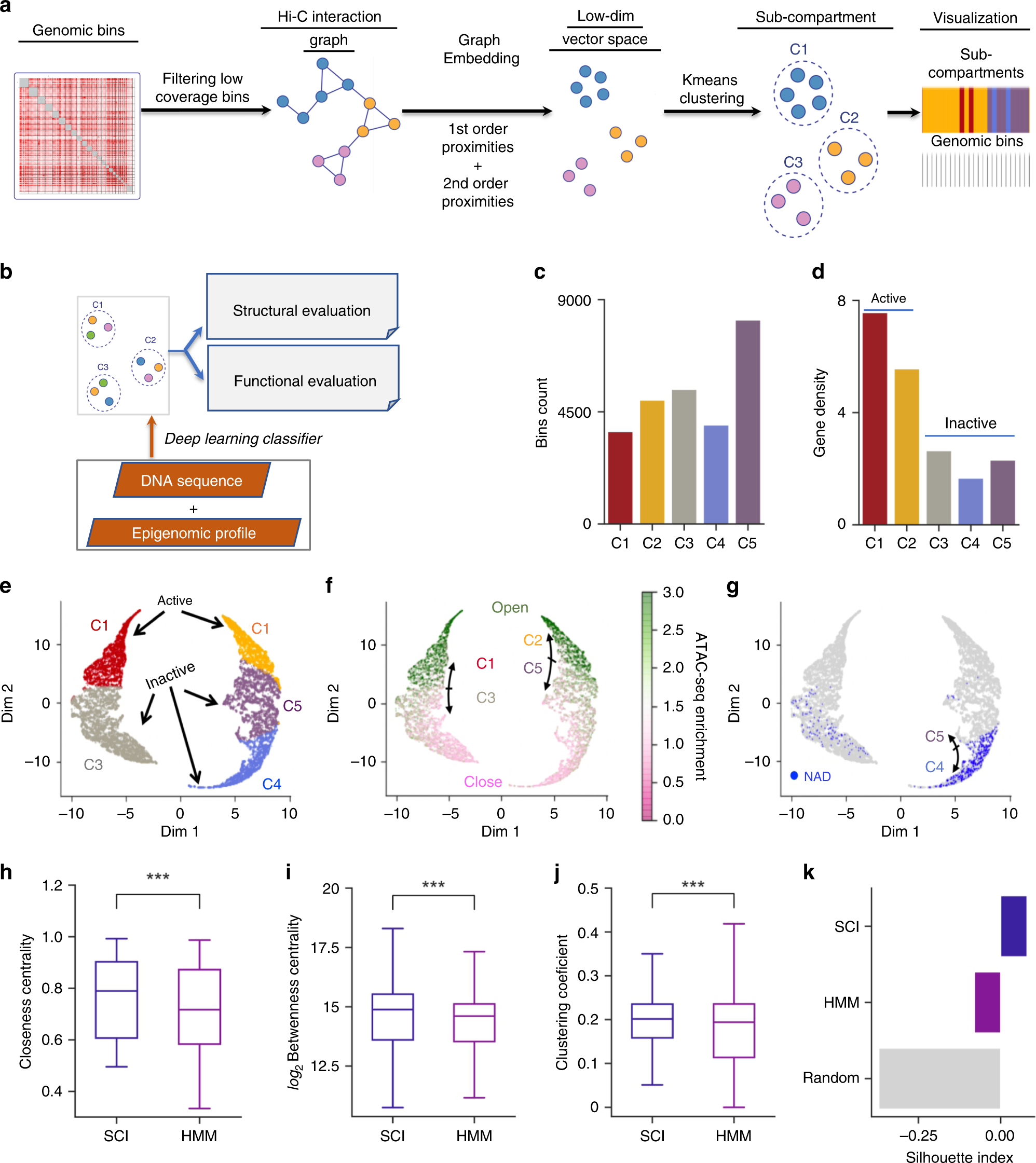
Graph embedding and unsupervised learning predict genomic sub
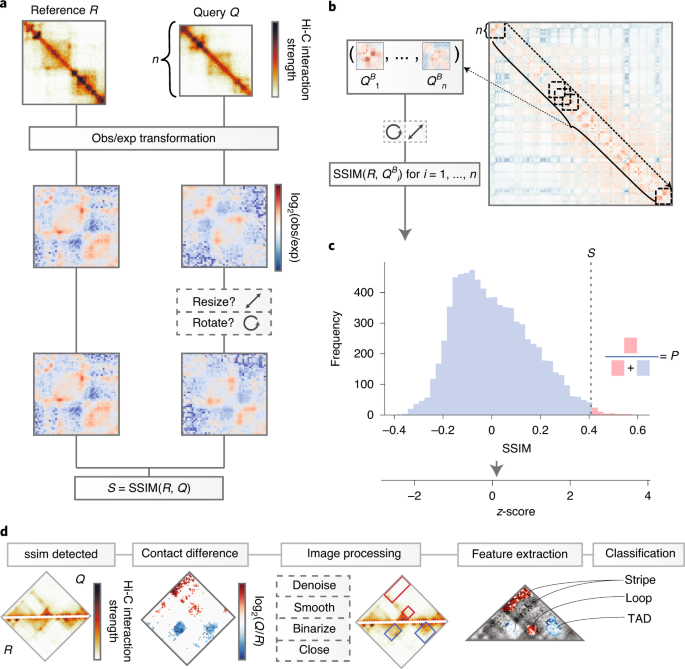
CHESS enables quantitative comparison of chromatin contact data

dcHiC: differential compartment analysis of Hi-C datasets
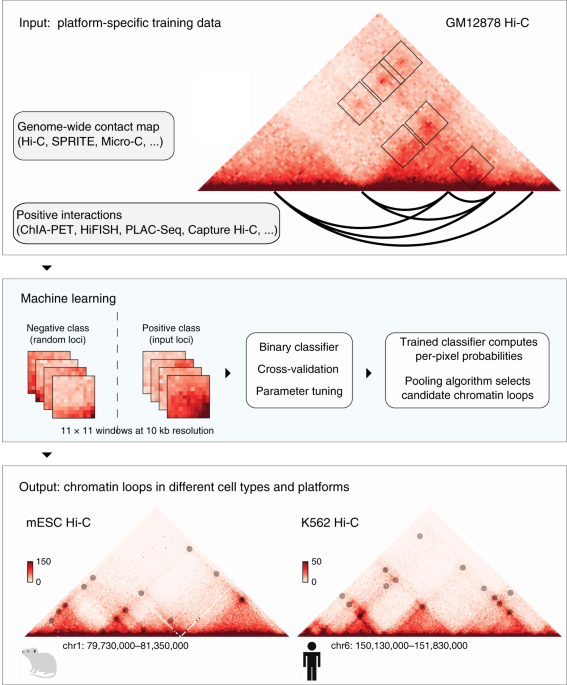
A supervised learning framework for chromatin loop detection in












